Gediting Technology
OUR CRISPR TOOLBOX
All of Akribion’s R&D efforts are based on a proprietary treasure box of more than 2,000 novel CRISPR nuclease sequences identified through a terabase-pair scale metagenomic screening. Based on this screening, two frontrunner nuclease families have been selected and optimized for their activity and IP position. We call these nucleases “G-dases®”.
G-dase E exhibits a novel RNA-dependent collateral molecular mechanism unique to CRISPR nucleases.
G-dase M is a more classical genome editing tool.
LEARN ABOUT HOW WE IDENTIFY G-DASES
Our
Discovery Process
To identify and engineer our novel G-dase® nucleases, we used metagenome sequencing techniques. Thus, we identified novel CRISPR-associated class 2 nucleases from metagenome samples that exhibit low sequence homology with other CRISPR nucleases and advanced them by protein engineering.
- Learning from nature: We selected metagenome samples by rational bioprospecting and isolated the DNA of all microorganisms from selected habitats.
- Metagenomics meets bioinformatics: We sequenced and analyzed the isolated DNA using state-of-the-art next-generation sequencing technologies to identify novel potential nucleases for genome editing.
- Finding new tools: We tested a wide variety of new genome editing nucleases to identify nucleases with high specificity and activity in different organisms of interest to build our G-dase® M nuclease family.
- Shaping the heroes: We optimized some selected metagenome sequences by protein engineering to enhance their activity and specificity. One of the best performing candidates then showed a surprising mode of action – this became our G-dase® E nuclease.
DISCOVER OUR G-DASES
G-dase M
Alternative Class II Type V CRISPR nuclease family for high precision genome editing in a broad range of organisms
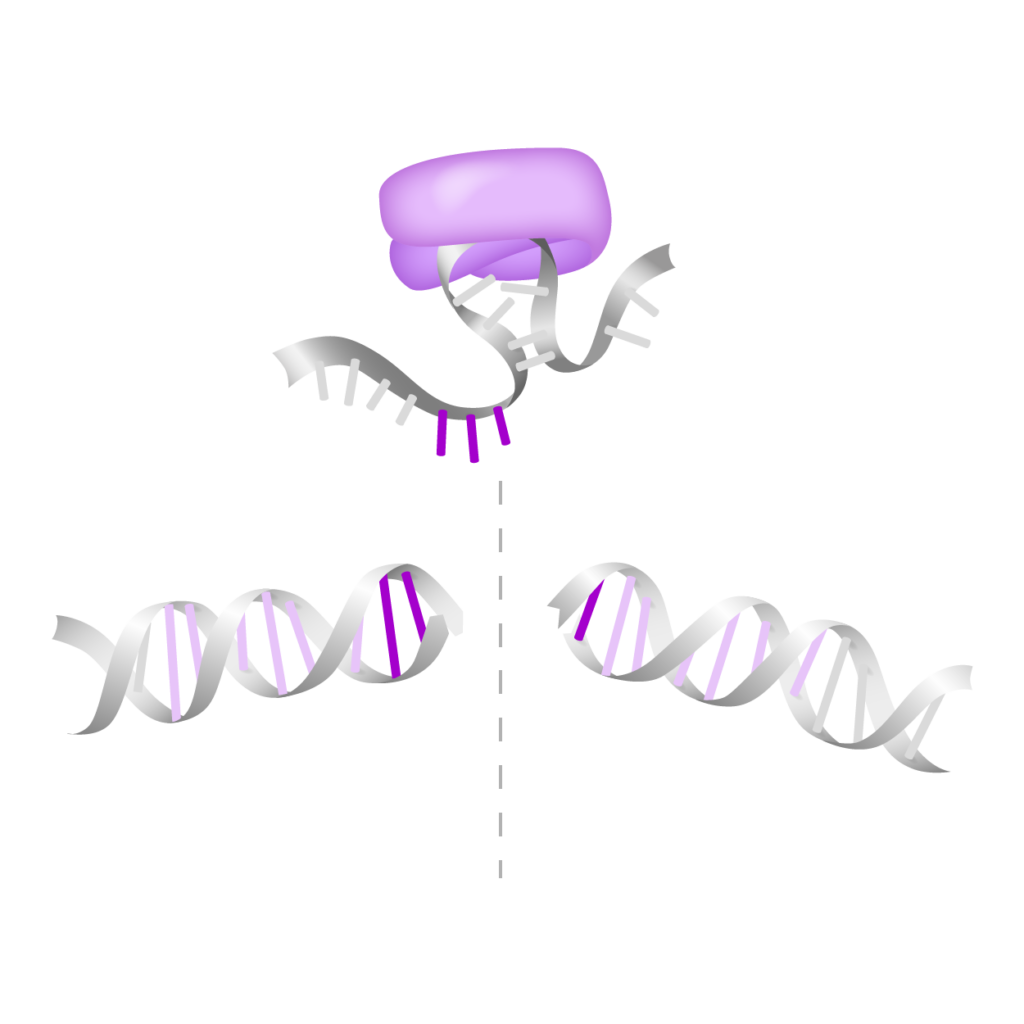

G-dase E
Engineered Class II Type V CRISPR nuclease with RNA-guided collateral DNA/RNAse activity for targeted cell depletion and HDR-based applications
USE OUR TECHNOLOGY